In this workshop we’ll learn how to build cloud-enabled web applications with React, AppSync, GraphQL, & AWS Amplify.
- GraphQL API with AWS AppSync
- Mocking and Testing
- Authentication
- Adding Authorization to the AWS AppSync API
- Lambda Resolvers
- Deploying the Services
- Hosting with the Amplify Console
- Amplify DataStore
- Deleting the resources
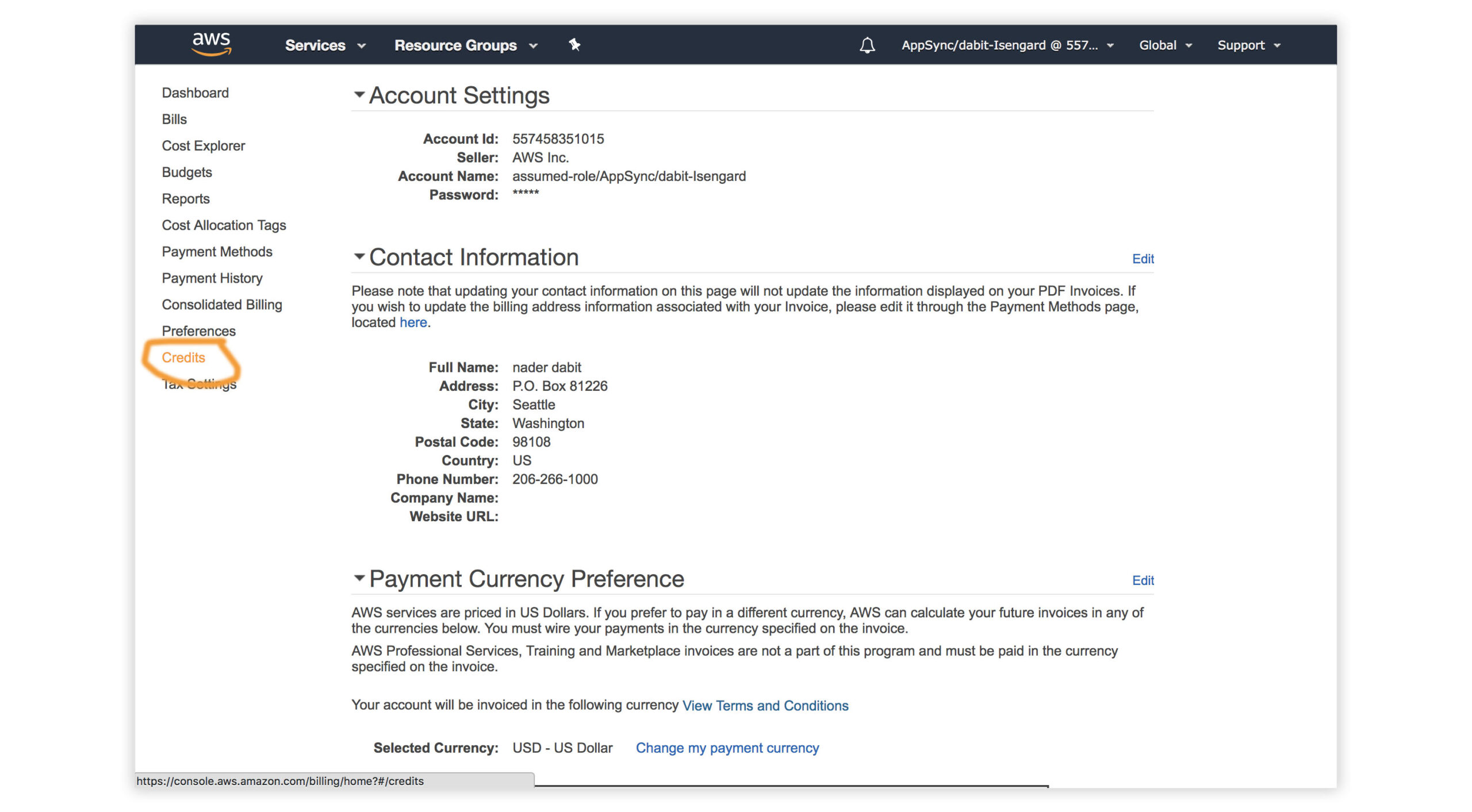
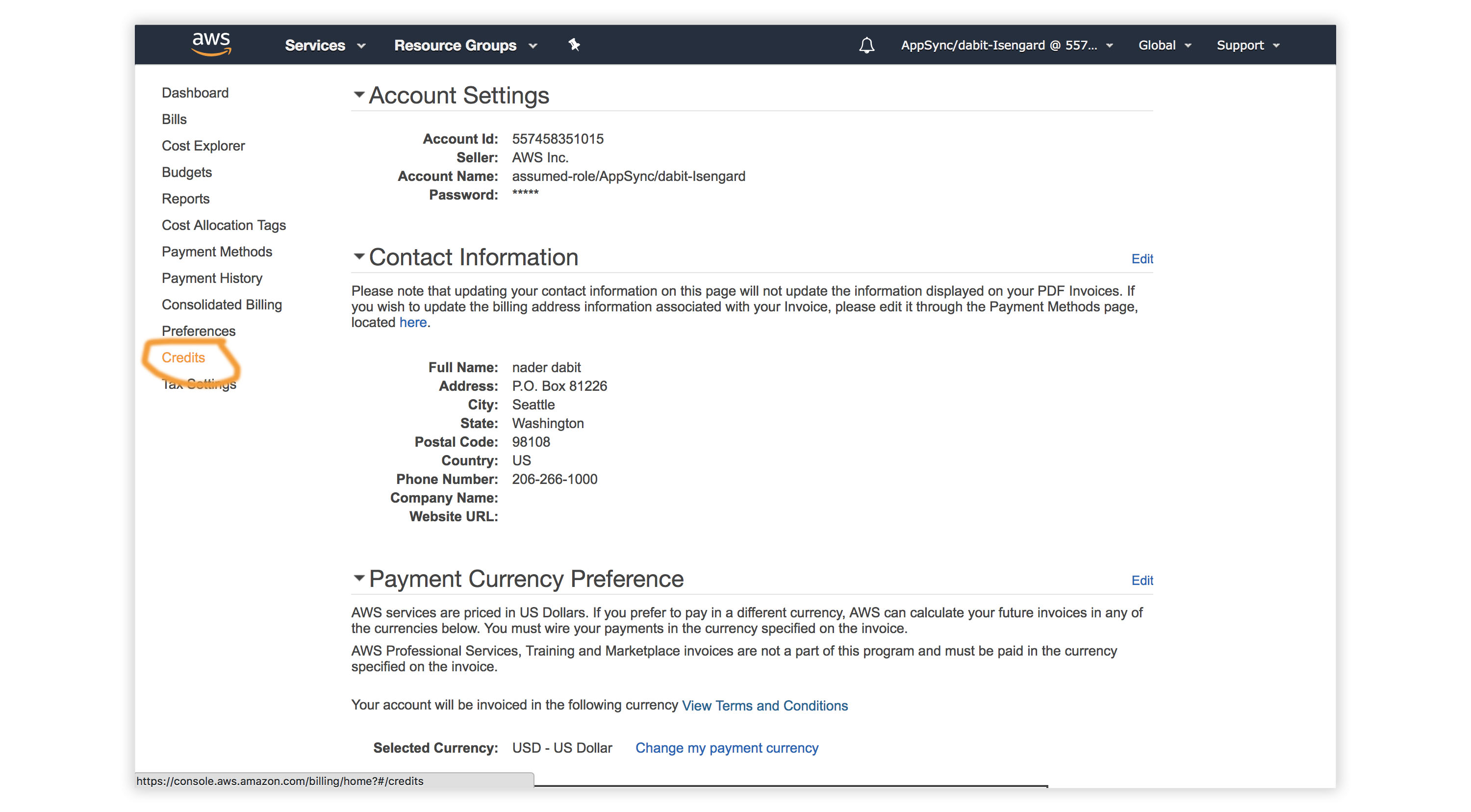
- Visit the AWS Console.
- In the top right corner, click on My Account.
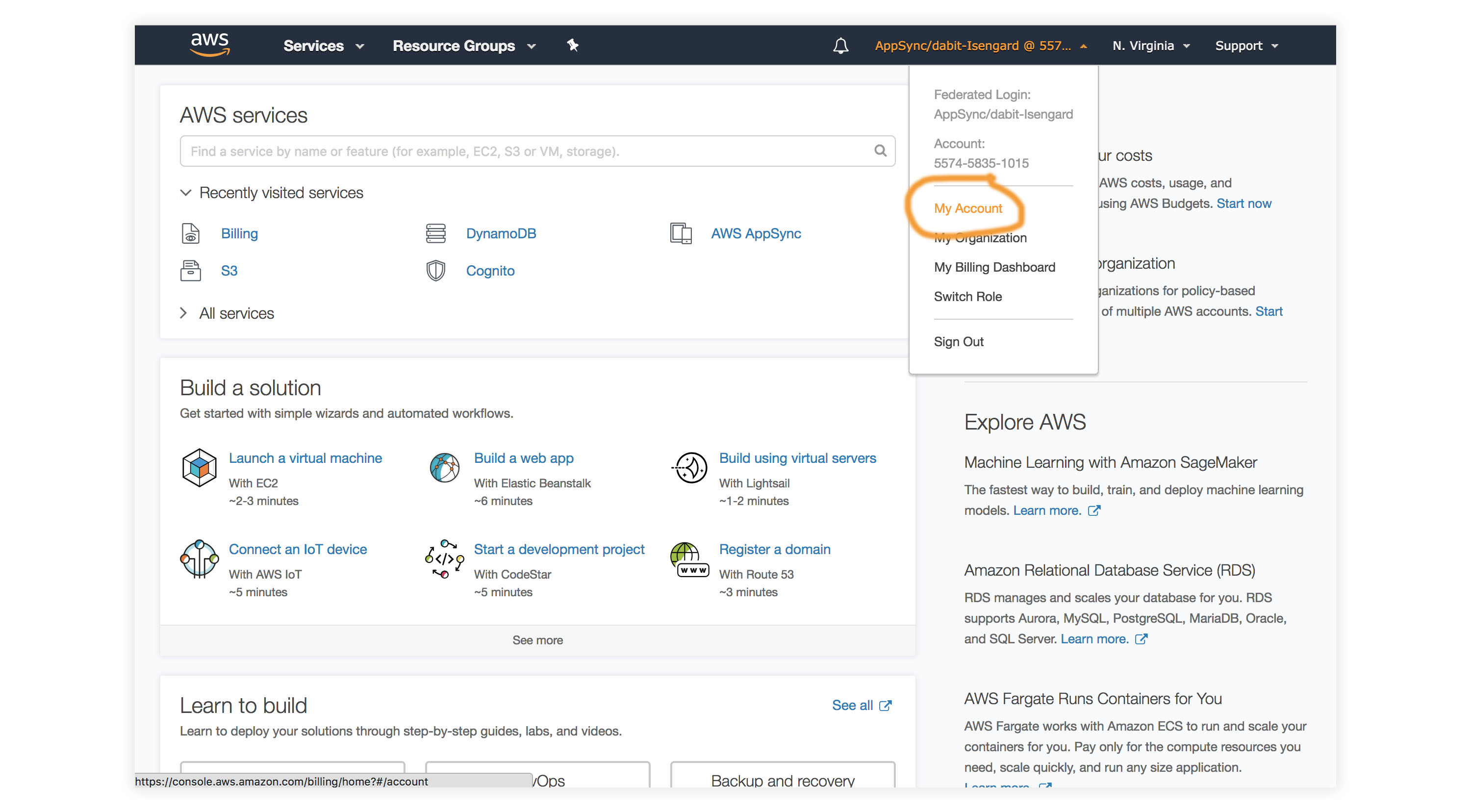
- In the left menu, click Credits.
To get started, we first need to create a new React project using the Create React App CLI.
$ npx create-react-app my-amplify-appNow change into the new app directory & install the AWS Amplify, AWS Amplify React, & uuid libraries:
$ cd my-amplify-app
$ npm install --save aws-amplify aws-amplify-react uuid
# or
$ yarn add aws-amplify aws-amplify-react uuidNext, we’ll install the AWS Amplify CLI:
$ npm install -g @aws-amplify/cliNow we need to configure the CLI with our credentials:
$ amplify configureIf you’d like to see a video walkthrough of this configuration process, click here.
Here we’ll walk through the amplify configure setup. Once you’ve signed in to the AWS console, continue:
- Specify the AWS Region: us-east-1 || us-west-2 || eu-central-1
- Specify the username of the new IAM user: amplify-workshop-user
In the AWS Console, click Next: Permissions, Next: Tags, Next: Review, & Create User to create the new IAM user. Then, return to the command line & press Enter.
- Enter the access key of the newly created user:
? accessKeyId: (<YOUR_ACCESS_KEY_ID>)
? secretAccessKey: (<YOUR_SECRET_ACCESS_KEY>) - Profile Name: amplify-workshop-user
$ amplify init- Enter a name for the project: amplifyreactapp
- Enter a name for the environment: dev
- Choose your default editor: Visual Studio Code (or your default editor)
- Please choose the type of app that you’re building javascript
- What javascript framework are you using react
- Source Directory Path: src
- Distribution Directory Path: build
- Build Command: npm run-script build
- Start Command: npm run-script start
- Do you want to use an AWS profile? Y
- Please choose the profile you want to use: amplify-workshop-user
Now, the AWS Amplify CLI has iniatilized a new project & you will see a new folder: amplify & a new file called aws-exports.js in the src directory. These files hold your project configuration.
To view the status of the amplify project at any time, you can run the Amplify status command:
$ amplify statusNow, our resources are created & we can start using them!
The first thing we need to do is to configure our React application to be aware of our new AWS Amplify project. We can do this by referencing the auto-generated aws-exports.js file that is now in our src folder.
To configure the app, open src/index.js and add the following code below the last import:
import Amplify from 'aws-amplify'
import config from './aws-exports'
Amplify.configure(config)Now, our app is ready to start using our AWS services.
To add a GraphQL API, we can use the following command:
$ amplify add api
? Please select from one of the above mentioned services: GraphQL
? Provide API name: ConferenceAPI
? Choose an authorization type for the API: API key
? Enter a description for the API key: <some description>
? After how many days from now the API key should expire (1-365): 365
? Do you want to configure advanced settings for the GraphQL API: No
? Do you have an annotated GraphQL schema? N
? Do you want a guided schema creation? Y
? What best describes your project: Single object with fields
? Do you want to edit the schema now? (Y/n) YWhen prompted, update the schema to the following:
# amplify/backend/api/ConferenceAPI/schema.graphql
type Talk @model {
id: ID!
clientId: ID
name: String!
description: String!
speakerName: String!
speakerBio: String!
}To mock and test the API locally, you can run the mock command:
$ amplify mock api
? Choose the code generation language target: javascript
? Enter the file name pattern of graphql queries, mutations and subscriptions: src/graphql/**/*.js
? Do you want to generate/update all possible GraphQL operations - queries, mutations and subscriptions: Y
? Enter maximum statement depth [increase from default if your schema is deeply nested]: 2This should start an AppSync Mock endpoint:
AppSync Mock endpoint is running at http://10.219.99.136:20002Open the endpoint in the browser to use the GraphiQL Editor.
From here, we can now test the API.
Execute the following mutation to create a new talk in the API:
mutation createTalk {
createTalk(input: {
name: "Full Stack React"
description: "Using React to build Full Stack Apps with GraphQL"
speakerName: "Jennifer"
speakerBio: "Software Engineer"
}) {
id name description speakerName speakerBio
}
}Now, let’s query for the talks:
query listTalks {
listTalks {
items {
id
name
description
speakerName
speakerBio
}
}
}We can even add search / filter capabilities when querying:
query listTalksWithFilter {
listTalks(filter: {
description: {
contains: "React"
}
}) {
items {
id
name
description
speakerName
speakerBio
}
}
}Now that the GraphQL API server is running we can begin interacting with it!
The first thing we’ll do is perform a query to fetch data from our API.
To do so, we need to define the query, execute the query, store the data in our state, then list the items in our UI.
// src/App.js
import React from 'react';
// imports from Amplify library
import { API, graphqlOperation } from 'aws-amplify'
// import query definition
import { listTalks as ListTalks } from './graphql/queries'
class App extends React.Component {
// define some state to hold the data returned from the API
state = {
talks: []
}
// execute the query in componentDidMount
async componentDidMount() {
try {
const talkData = await API.graphql(graphqlOperation(ListTalks))
console.log('talkData:', talkData)
this.setState({
talks: talkData.data.listTalks.items
})
} catch (err) {
console.log('error fetching talks...', err)
}
}
render() {
return (
<>
{
this.state.talks.map((talk, index) => (
<div key={index}>
<h3>{talk.speakerName}</h3>
<h5>{talk.name}</h5>
<p>{talk.description}</p>
</div>
))
}
</>
)
}
}
export default AppIn the above code we are using API.graphql to call the GraphQL API, and then taking the result from that API call and storing the data in our state. This should be the list of talks you created via the GraphiQL editor.
Next, test the app locally:
$ npm startNow, let’s look at how we can create mutations.
To do so, we’ll refactor our initial state in order to also hold our form fields and add an event handler.
We’ll also be using the API class from amplify again, but now will be passing a second argument to graphqlOperation in order to pass in variables: API.graphql(graphqlOperation(CreateTalk, { input: talk })).
We also have state to work with the form inputs, for name, description, speakerName, and speakerBio.
// src/App.js
import React from 'react';
import { API, graphqlOperation } from 'aws-amplify'
// import uuid to create a unique client ID
import uuid from 'uuid/v4'
import { listTalks as ListTalks } from './graphql/queries'
// import the mutation
import { createTalk as CreateTalk } from './graphql/mutations'
const CLIENT_ID = uuid()
class App extends React.Component {
// define some state to hold the data returned from the API
state = {
name: '', description: '', speakerName: '', speakerBio: '', talks: []
}
// execute the query in componentDidMount
async componentDidMount() {
try {
const talkData = await API.graphql(graphqlOperation(ListTalks))
console.log('talkData:', talkData)
this.setState({
talks: talkData.data.listTalks.items
})
} catch (err) {
console.log('error fetching talks...', err)
}
}
createTalk = async() => {
const { name, description, speakerBio, speakerName } = this.state
if (name === '' || description === '' || speakerBio === '' || speakerName === '') return
const talk = { name, description, speakerBio, speakerName, clientId: CLIENT_ID }
const talks = [...this.state.talks, talk]
this.setState({
talks, name: '', description: '', speakerName: '', speakerBio: ''
})
try {
await API.graphql(graphqlOperation(CreateTalk, { input: talk }))
console.log('item created!')
} catch (err) {
console.log('error creating talk...', err)
}
}
onChange = (event) => {
this.setState({
[event.target.name]: event.target.value
})
}
render() {
return (
<>
<input
name='name'
onChange={this.onChange}
value={this.state.name}
placeholder='name'
/>
<input
name='description'
onChange={this.onChange}
value={this.state.description}
placeholder='description'
/>
<input
name='speakerName'
onChange={this.onChange}
value={this.state.speakerName}
placeholder='speakerName'
/>
<input
name='speakerBio'
onChange={this.onChange}
value={this.state.speakerBio}
placeholder='speakerBio'
/>
<button onClick={this.createTalk}>Create Talk</button>
{
this.state.talks.map((talk, index) => (
<div key={index}>
<h3>{talk.speakerName}</h3>
<h5>{talk.name}</h5>
<p>{talk.description}</p>
</div>
))
}
</>
)
}
}
export default AppNext, let’s update the app to add authentication.
To add authentication, we can use the following command:
$ amplify add auth
? Do you want to use default authentication and security configuration? Default configuration
? How do you want users to be able to sign in when using your Cognito User Pool? Username
? Do you want to configure advanced settings? No, I am done. To add authentication in the React app, we’ll go into src/App.js and first import the withAuthenticator HOC (Higher Order Component) from aws-amplify-react:
// src/App.js, import the new component
import { withAuthenticator } from 'aws-amplify-react'Next, we’ll wrap our default export (the App component) with the withAuthenticator HOC:
// src/App.js, change the default export to this:
export default withAuthenticator(App, { includeGreetings: true })To deploy the authentication service and mock and test the app locally, you can run the mock command:
$ amplify mock
? Are you sure you want to continue? YesNext, to test it out in the browser:
npm startNow, we can run the app and see that an Authentication flow has been added in front of our App component. This flow gives users the ability to sign up & sign in.
We can access the user’s info now that they are signed in by calling Auth.currentAuthenticatedUser() in componentDidMount.
import {API, graphqlOperation, /* new 👉 */ Auth} from 'aws-amplify'
async componentDidMount() {
// add this code to componentDidMount
const user = await Auth.currentAuthenticatedUser()
console.log('user:', user)
console.log('user info:', user.signInUserSession.idToken.payload)
}Next we need to update the AppSync API to now use the newly created Cognito Authentication service as the authentication type.
To do so, we’ll reconfigure the API:
$ amplify update api
? Please select from one of the below mentioned services: GraphQL
? Choose the default authorization type for the API: Amazon Cognito User Pool
? Do you want to configure advanced settings for the GraphQL API: No, I am doneNext, we’ll test out the API with authentication enabled:
$ amplify mockNow, we can only access the API with a logged in user.
You’ll notice an auth button in the GraphiQL explorer that will allow you to update the simulated user and their groups.
For authorization rules, we can start using the @auth directive.
What if you’d like to have a new Comment type that could only be updated or deleted by the creator of the Comment but can be read by anyone?
We could add the following type to our GraphQL schema:
# amplify/backend/api/ConferenceAPI/schema.graphql
type Comment @model @auth(rules: [
{ allow: owner, ownerField: "createdBy", operations: [create, update, delete]},
{ allow: private, operations: [read] }
]) {
id: ID!
message: String
createdBy: String
}allow: owner – This allows us to set owner authorization rules.
allow: private – This allows us to set private authorization rules.
This would allow us to create comments that only the creator of the Comment could delete, but anyone could read.
Creating a comment:
mutation createComment {
createComment(input:{
message: "Cool talk"
}) {
id
message
createdBy
}
}Listing comments:
query listComments {
listComments {
items {
id
message
createdBy
}
}
}Updating a comment:
mutation updateComment {
updateComment(input: {
id: "59d202f8-bfc8-4629-b5c2-bdb8f121444a"
}) {
id
message
createdBy
}
}If you try to update a comment from someone else, you will get an unauthorized error.
What if we wanted to create a relationship between the Comment and the Talk? That’s pretty easy. We can use the @connection directive:
# amplify/backend/api/ConferenceAPI/schema.graphql
type Talk @model {
id: ID!
clientId: ID
name: String!
description: String!
speakerName: String!
speakerBio: String!
comments: [Comment] @connection(name: "TalkComments")
}
type Comment @model @auth(rules: [
{ allow: owner, ownerField: "createdBy", operations: [create, update, delete]},
{ allow: private, operations: [read] }
]) {
id: ID!
message: String
createdBy: String
talk: Talk @connection(name: "TalkComments")
}Because we’re updating the way our database is configured by adding relationships which requires a global secondary index, we need to delete the old local database:
$ rm -r amplify/mock-dataNow, restart the server:
$ amplify mockNow, we can create relationships between talks and comments. Let’s test this out with the following operations:
mutation createTalk {
createTalk(input: {
id: "test-id-talk-1"
name: "Talk 1"
description: "Cool talk"
speakerBio: "Cool gal"
speakerName: "Jennifer"
}) {
id
name
description
}
}
mutation createComment {
createComment(input: {
commentTalkId: "test-id-talk-1"
message: "Great talk"
}) {
id message
}
}
query listTalks {
listTalks {
items {
id
name
description
comments {
items {
message
createdBy
}
}
}
}
}If you’d like to read more about the @auth directive, check out the documentation here.
The last problem we are facing is that anyone signed in can create a new talk. Let’s add authorization that only allows users that are in an Admin group to create and update talks.
# amplify/backend/api/ConferenceAPI/schema.graphql
type Talk @model @auth(rules: [
{ allow: groups, groups: ["Admin"] },
{ allow: private, operations: [read] }
]) {
id: ID!
clientId: ID
name: String!
description: String!
speakerName: String!
speakerBio: String!
comments: [Comment] @connection(name: "TalkComments")
}
type Comment @model @auth(rules: [
{ allow: owner, ownerField: "createdBy", operations: [create, update, delete]},
{ allow: private, operations: [read] }
]) {
id: ID!
message: String
createdBy: String
talk: Talk @connection(name: "TalkComments")
}Run the server:
$ amplify mockClick on the auth button and add Admin the user’s groups.
Now, you’ll notice that only users in the Admin group can create, update, or delete a talk, but anyone can read it.
Next, let’s have a look at how to deploy a serverless function and use it as a GraphQL resolver.
The use case we will work with is fetching data from another HTTP API and returning the response via GraphQL. To do this, we’ll use a serverless function.
The API we will be working with is the CoinLore API that will allow us to query for cryptocurrency data.
To get started, we’ll create the new function:
$ amplify add function
? Provide a friendly name for your resource to be used as a label for this category in the project: currencyfunction
? Provide the AWS Lambda function name: currencyfunction
? Choose the function template that you want to use: Hello world function
? Do you want to access other resources created in this project from your Lambda function? N
? Do you want to edit the local lambda function now? YUpdate the function with the following code:
// amplify/backend/function/currencyfunction/src/index.js
const axios = require('axios')
exports.handler = function (event, _, callback) {
let apiUrl = `https://api.coinlore.com/api/tickers/?start=1&limit=10`
if (event.arguments) {
const { start = 0, limit = 10 } = event.arguments
apiUrl = `https://api.coinlore.com/api/tickers/?start=${start}&limit=${limit}`
}
axios.get(apiUrl)
.then(response => callback(null, response.data.data))
.catch(err => callback(err))
}In the above function we’ve used the axios library to call another API. In order to use axios, we need be sure that it will be installed by updating the package.json for the new function:
amplify/backend/function/currencyfunction/src/package.json
"dependencies": {
// ...
"axios": "^0.19.0",
},Next, we’ll update the GraphQL schema to add a new type and query. In amplify/backend/api/ConferenceAPI/schema.graphql, update the schema with the following new types:
type Coin {
id: String!
name: String!
symbol: String!
price_usd: String!
}
type Query {
getCoins(limit: Int start: Int): [Coin] @function(name: "currencyfunction-${env}")
}Now the schema has been updated and the Lambda function has been created. To test it out, you can run the mock command:
$ amplify mockIn the query editor, run the following queries:
# basic request
query listCoins {
getCoins {
price_usd
name
id
symbol
}
}
# request with arguments
query listCoinsWithArgs {
getCoins(limit:3 start: 10) {
price_usd
name
id
symbol
}
}This query should return an array of cryptocurrency information.
Next, let’s deploy the AppSync GraphQL API and the Lambda function:
$ amplify push
? Do you want to generate code for your newly created GraphQL API? Y
? Choose the code generation language target: javascript
? Enter the file name pattern of graphql queries, mutations and subscriptions: src/graphql/**/*.js
? Do you want to generate/update all possible GraphQL operations - queries, mutations and subscriptions? Y
? Enter maximum statement depth [increase from default if your schema is deeply nested] 2To view the new AWS AppSync API at any time after its creation, run the following command:
$ amplify console apiTo view the Cognito User Pool at any time after its creation, run the following command:
$ amplify console authTo test an authenticated API out in the AWS AppSync console, it will ask for you to Login with User Pools. The form will ask you for a ClientId. This ClientId is located in src/aws-exports.js in the aws_user_pools_web_client_id field.
The Amplify Console is a hosting service with continuous integration and continuous deployment.
The first thing we need to do is create a new GitHub repo for this project. Once we’ve created the repo, we’ll copy the URL for the project to the clipboard & initialize git in our local project:
$ git init
$ git remote add origin git@github.com:username/project-name.git
$ git add .
$ git commit -m 'initial commit'
$ git push origin masterNext we’ll visit the Amplify Console in our AWS account at https://us-east-1.console.aws.amazon.com/amplify/home.
Here, we’ll click on the app that we deployed earlier.
Next, under “Frontend environments”, authorize Github as the repository service.
Next, we’ll choose the new repository & branch for the project we just created & click Next.
In the next screen, we’ll create a new role & use this role to allow the Amplify Console to deploy these resources & click Next.
Finally, we can click Save and Deploy to deploy our application!
Now, we can push updates to Master to update our application.
To implement a GraphQL API with Amplify DataStore, check out the tutorial here
If at any time, or at the end of this workshop, you would like to delete a service from your project & your account, you can do this by running the amplify remove command:
$ amplify remove auth
$ amplify pushIf you are unsure of what services you have enabled at any time, you can run the amplify status command:
$ amplify statusamplify status will give you the list of resources that are currently enabled in your app.
If you’d like to delete the entire project, you can run the delete command:
$ amplify delete